Purines are vital organic compounds that play a fundamental role in various biological processes, such as the synthesis of DNA and RNA, energy metabolism, and cell signaling. These compounds are essential building blocks in the production of nucleotides, which are the fundamental units of genetic material.
Purine biosynthesis is a highly regulated process responsible for synthesizing purines from simple precursors, such as amino acids and formate.
The production of these precursors requires a great deal of energy, so regulating purine biosynthesis is critical to ensure that cells only produce the necessary amounts of purines at the right time.
Additionally, purine biosynthesis is essential in producing essential signaling molecules such as cyclic AMP and GTP, which play key roles in cellular metabolism and signaling.
Understanding purine biosynthesis and the regulation of purine metabolism is critical for maintaining the body’s purine balance and preventing disorders such as gout, Lesch-Nyhan syndrome, and various immunodeficiency disorders. Therefore, research on purine biosynthesis is of significant importance for basic research and clinical medicine.
In 1948, “John Buchanor” obtained the first clues about how this process occurs. Denovo by feeding a variety of isotopically labeled compounds to pigeons and chemically determining the position of the labeled atoms in their excreted uric acid.
AMP and GMP are nucleic acid purine nucleotides that contain the purine bases Adenine (A) and Guanine (G). The results of the studies concern purine synthesis.
What is purine synthesis?
Purine synthesis is the process by which purine nucleotides, such as adenosine and guanosine, are synthesized de novo within the body.
- Purine nucleotides are essential building blocks for DNA and RNA and critical components of various metabolic pathways and signaling molecules.
- The synthesis of purine nucleotides occurs through a series of enzymatic reactions that convert simpler precursors into final products, including converting amino acids and other metabolites into nucleotide bases.
- Purine synthesis is a tightly regulated process essential for maintaining the proper balance of nucleotides in the body and is crucial for various biological processes such as DNA replication, cell division, and energy metabolism.
- Dysregulation of purine synthesis can lead to multiple disorders, including gout and Lesch-Nyhan syndrome.
Sources of atoms in Purine Ring
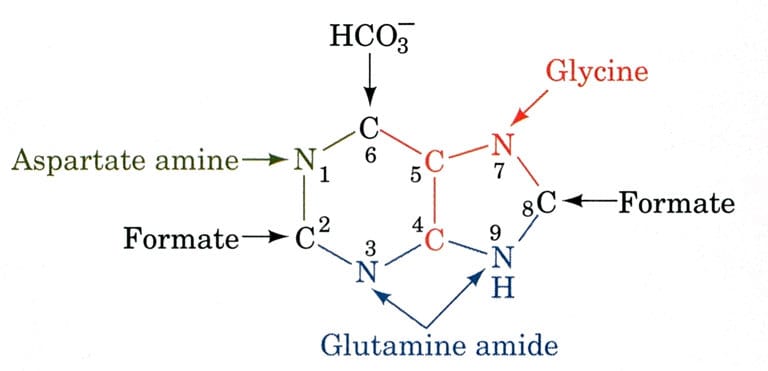
The biosynthetic organs of purine ring atoms note that C4, C5, and N7 come from a single glycine molecule, but each of the other atoms is derived from an independent precursor.
- N1→ came from Asparagine
- C2 and C8 → came from formate (THF)
- N3 and N9 → came from Glutamine
- C4, C5, and N7 → came from HCO3–
Purine Synthesis Pathways
Purine biosynthesis pathway can be explained in two different ways
- De-Novo Pathway
- Salvage Pathway (also called Dust-bin Pathway)
De Novo Purine Synthesis
In this de novo synthesis of purines, each atom in the purine nucleotide came from different sources mentioned in the structure and data. There are three significant steps involved in this purine synthesis pathway. There are 10 steps of purine synthesis. Let us take the purine synthesis steps one by one.
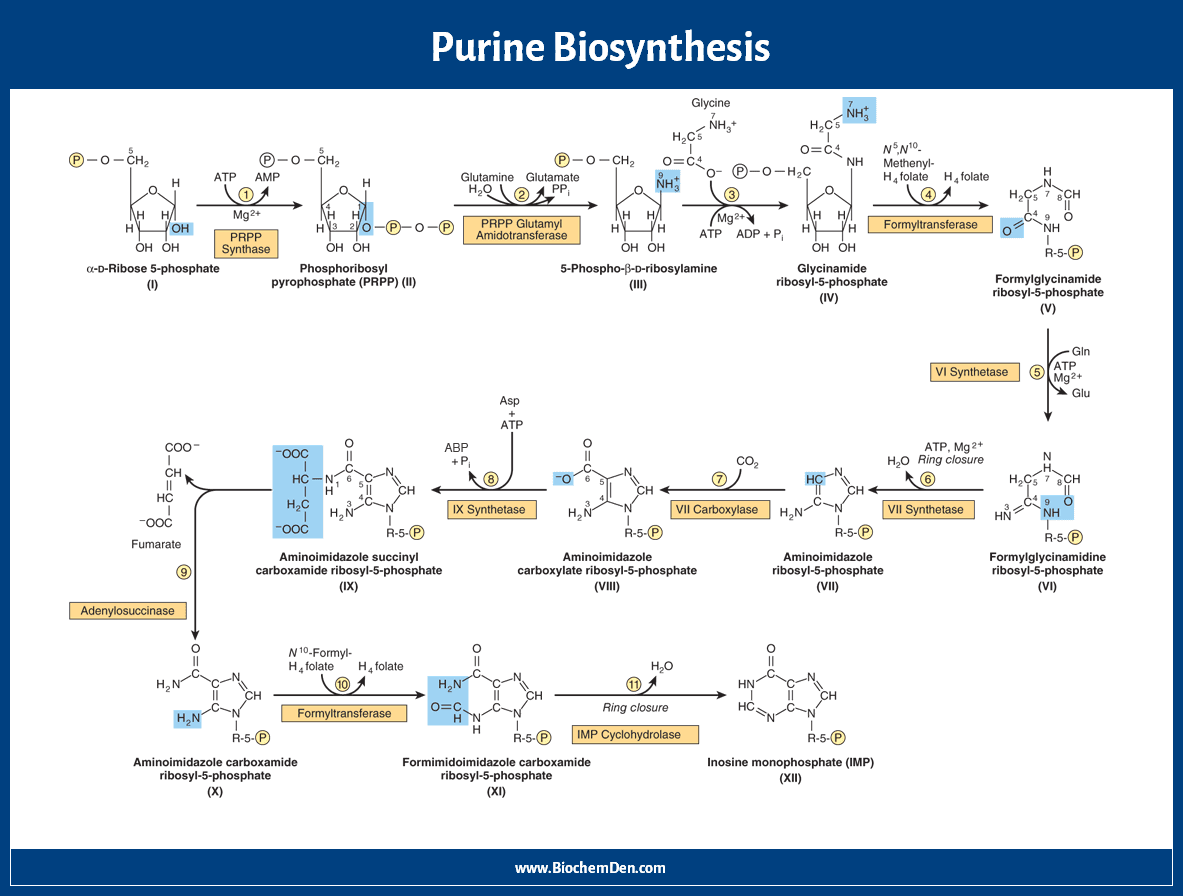
- Ribose-5-Phosphate to IMP synthesis
- Synthesis of AMP from IMP
- Synthesis of GMP from IMP
Ribose-5-Phosphate to IMP synthesis
Here are the steps of de novo synthesis of purine nucleotides.
Step 1: Amination
The starting material for purine biosynthesis is ribose-5-P, a product of the Hexose Monophosphate Shunt or Pentose Phosphate Pathway (HMP Shunt). In this reaction, ribose-5-phosphate is converted into phosphoribosyl pyrophosphate by pyrophosphokinase, and ATP is consumed.
PRPP + Glutamine + H2O → PRA + Glutamate + PPi
(PRPP = 5-phosphoribosyl-1-pyrophosphate, PRA = 5-phosphoribosyl-1-amine)
Step 2: Addition of N9
One nitrogen is added on Ribose-5-P, to form 5-phosphoribosyl-1-amine (PRA). The nitrogen is donated by Glutamine. Ribose-5-Phosphate is derived from PRPP.
The reaction needs energy from ATP hydrolysis. This is the rate-limiting enzyme for this pathway. This step is inhibited by azaserine, the anticancer drug.
Step 3: Incorporation of C4, C5, and N7
When phosphoribosyl amine (PRA) is condensed with glycine, glycinamide ribotide (GAR) is formed. This reaction is catalyzed by GAR synthase.
PRA + ATP → GAR + PPi
Step 4: Addition of C8
Formyl glycine amide ribotide (FGAR) is formed from glycinamide ribotide. This reaction is catalyzed by transformylase. The Formyl donor in this case is N10-Formyl-THF.
GAR + FGAM → FGAR + AICAR
Step 5: Addition of N3
Formyl Glycine ribotide is converted into Formylglycinaidine ribotide (FGAM) in the presence of the enzyme FGAM synthetase. Here amide donor is Glutamine and it is ATP consumed reaction. This enzyme is also inhibited by azaserine.
FGAR + ATP → FGAM + ADP
Step 6: Cyclisation (Closure of Ring)
FGAM is converted into 5-amino imidazole ribotide (AIR). This reaction is catalyzed by AIR synthetase. Here, ATP is consumed.
FGAM → AIR + NH3
Step 7: Addition of C6
Amino imidazole ribotide is converted into 5-amino-4-carboxy-amino-imidazole ribonucleotide (CAIR). This reaction is catalyzed by AIR carboxylase. In this reaction, carbonic acid is substituted with a 4th carbon atom in the form of the carboxyl group (CAIR).
AIR → CAIR + H2O
This carbon dioxide fixation reaction does not require biotin or ATP.
Step 8: Addition of N1
Carboxy Amino Imidazole has converted into 5-AminoImidazole (N-Succinylocarboxamide) ribotide (SACAIR). This reaction is catalyzed by SACAIR synthetase. In this reaction, one Aspartic acid linked with Carboxyl group ATP is consumed.
CAIR → SAICAR + CO2
Step 9: Removal of Fumaric acid
SACAIR is converted into 5-AminoImidazole-4-CarboxyAmide Ribotide (AICAR). This reaction is catalyzed by adenosuccinate lyase. The linked aspartic acid is hydrolyzed into fumarate, which directly enters the TCA cycle.
The amino group of aspartic acid becomes the first nitrogen of the purine ring.
Step 10: Addition of C2
AICAR is converted into 5-Formamino-Imidazole-4-Carboxamide Ribotide (FAICAR). This reaction is catalyzed by transformylase.
Here formyl group donor is N10-Formyl THF. This carbon is derived from the one-carbon pool.
In folic acid deficiency, this step is blocked; hence orange-coloured FAICAR is excreted in the urine.
Step 11: Cyclization
FAICAR is converted into Inosine Mono Phosphate (IMP) by the catalyzation process. This reaction is catalyzed by IMP Cyclohydrolase. It contains purine, hypoxanthine.
SAICAR → IMP + Fumarate
| Donor atom | Added features | Special | Product |
| Glutamine | N9 | Rate-limiting | PRA |
| Glycine | C4, C5 and N7 | ATP required | GAR |
| Methenyl-THFA | C8 | – | FGAR |
| Glutamine | N3 | ATP required | FGAM |
| – | Ring closure | ATP need | AIR |
| Carbon dioxide | C6 | – | ACAIR |
| Aspartic acid | N1 | ATP required | SAICAR |
| – | – | Fumarate removed | AICAR |
| Formyl THFA | C2 | – | FAICAR |
| – | – | Ring closure | IMP |
Synthesis of AMP and GMP from IMP
Adenosine monophosphate (AMP) and guanosine monophosphate (GMP) are essential nucleotides that play a crucial role in various biological processes.
- AMP and GMP are synthesized from the precursor inosine monophosphate (IMP) via different pathways.
- The synthesis of AMP from IMP is carried out through a series of reactions involving adding an amino group. In contrast, the synthesis of GMP from IMP consists of adding a formyl group.
- Understanding the mechanisms behind synthesizing these nucleotides is essential for gaining insight into their biological functions and the potential therapeutic applications of targeting their biosynthesis.
Synthesis of AMP from IMP
IMP is the central intermediate of both AMP and UMP.
Step 1: IMP to AdenyloSuccinate
The IMP is converted into adenyloSuccinate by taking Aspartate and GTP, which gives the power of the UTP to GTP and inorganic phosphate. This reaction is catalyzed by adenyl-succinate synthetase.
Step 2: AdenyloSuccinate to AMP
AdenyloSuccinate is converted into AMP by releasing Aspartate in the form of fumarate. This reaction is catalyzed by Adenylo Succinate Lyase.
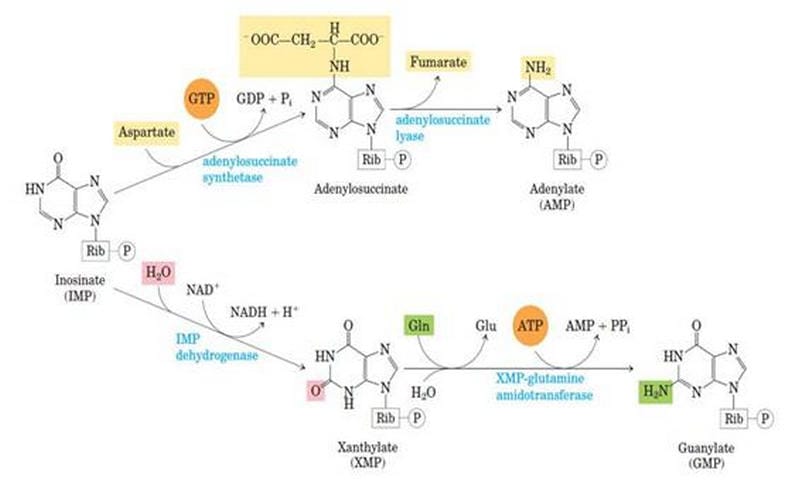
Synthesis of GMP from IMP
Step 1: IMP to XMP
In the presence of the enzyme IMP-dehydrogenase, IMP is converted into Xanthosine Monophosphate. This is called dehydrogenation.
Step 2: XMP to GMP
XMP is converted into GMP by the enzyme GMP synthase. Here, the amino group donor is glutamate.
Overall equation of Purine Synthesis
PRPP + Glutamine + ATP → IMP + PPi + Glutamate + Fumarate + CO2 + NH3 + 2ADP
Regulation of Purine Biosynthesis
Activation:
- The activator molecule for purine synthesis is PRPP, which activates the enzyme AmidoPhospho Ribosyl transferase
- The initiator molecule for the synthesis is ribose-5-phosphate. The optimum concentration of Ribos-5-Phosphate is maintained by the enzyme Glucose-6-Phosphate dehydrogenase, which is the regulatory enzyme of Hexose Mono Phosphate Shunt.
Inhibition:
- The rate-limiting enzyme “Ribose-5-Phosphate Pyrophospho kinase” inhibited the nucleotides AMP, ADP, ATP, GMP, GDP, and GTP by a feedback inhibition mechanism.
- The enzyme amino phosphoribosyl transferase is inhibited by AMP, ADP, ATP, GMP, GDP, GTP adenylosuccinate and XMP.
- The amp inhibits Adenylo succinate synthatase GMP inhibits IMP dehydrogenase.
Salvage pathway of purine synthesis
This is another type of purine nucleotide synthesis from scratch. So, this is also called the “Dust-bin pathway”. Most cells have an active turnover of many of their nucleic acids, resulting in adenine, guanine, and Hypoxanthine.
Purines that result from the normal turnover of cellular nucleic acids or that are obtained from the diet are not degraded. It can be reconverted into nucleoside triphosphate and used by the body. This is referred to as the “salvage pathway” for purines.
Two enzymes are involved in this pathway.
APRT Enzyme
- APRT means Adenosyl Phosphoribosyl Transferase (APRTase)
- This enzyme catalyzes the reaction of Adenine to AMP conversion. In this reaction the secondary substrate is PRPP and byproduct is PPi.
Adenine + PRPP ↔ AMP + PPi
HGPRT Enzyme
- HGPRT means Hypoxanthine-Guanine Phospho Ribosyl Transferase (HGPRTase).
- This enzyme catalyzes the reaction of GMP formation from Hypoxanthine and PRPP.
Hypoxanthine + PRPP ↔ IMP + PPi
Guanine + PRPP ↔ GMP + PPi
The salvage pathway is of special importance in tissues like RBC and the brain, where the de-novo pathway is not operating.
The origin of the carbon and nitrogen atoms of the purine ring system was determined by John Buchanan using an isotopic tracer experiment.
The detailed explanation of purine biosynthesis was first explained by Buchanan and G.Robert Greenberg in the 1950s.
Disorders of Purine Metabolism
Here’s the content for the “Disorders of Purine Metabolism” section in a point-wise format:
- Disorders of purine metabolism refer to a group of inherited metabolic disorders that affect the body’s ability to process purines.
- Gout is a common purine metabolism disorder that results from the deposition of uric acid crystals in joints and other tissues, causing inflammation and pain.
- Lesch-Nyhan syndrome is a rare disorder that affects the metabolism of purines and results in hyperuricemia, neurological symptoms, and self-injurious behaviour.
- Other disorders of purine metabolism include adenine phosphoribosyltransferase deficiency, purine nucleoside phosphorylase deficiency, and hypoxanthine-guanine phosphoribosyltransferase deficiency (HGPRT).
- These disorders can lead to various symptoms, including joint pain, muscle weakness, intellectual disability, neurological symptoms, and behavioral problems.
- The treatment of purine metabolism disorders often involves a combination of medication and lifestyle changes, such as dietary modifications to reduce purine intake and increase hydration.
- In some cases, enzyme replacement therapy or stem cell transplantation is a treatment option.
- Ongoing research is focused on developing new treatments for purine metabolism disorders and gaining a better understanding of their underlying mechanisms.
- Advances in genetic testing have also allowed for early diagnosis and management of purine metabolism disorders, leading to better outcomes for patients.
Frequently Asked Questions (FAQs)
How is purine synthesized?
Pyrimidine synthesis begins with the binding of a carbon atom to one of the five nitrogen atoms in cytosine (C). This process is catalyzed by a purine nucleobase cyclase, which uses ATP as an energy source. The next step in pyrimidine synthesis is the attachment of a uracil group to the phosphate group on the pyrimidine base.
Where is purine synthesis?
Purine synthesis is the process of producing purines, a group of organic molecules that includes three important molecules: adenine, guanine, and thymine. Purines are used in DNA and RNA.
What is the purpose of purine synthesis?
Purine synthesis is the process by which cells produce adenosine triphosphate (ATP), a molecule that provides energy for cellular metabolism.
Why is purine important in DNA synthesis?
Pyrimidine bases are important in DNA synthesis because they are incorporated into the DNA double helix during the process of replication.
What are purines?
Pyrimidines are a class of molecules that contain one or more nitrogen atoms. Purines are found in many foods and can be converted into uric acid.
How do you remember the purine synthesis?
The purine synthesis starts with the conversion of methylamine to guanidine by the enzyme methylamine dehydrogenase. Guanidine is then converted to urea by the enzyme urea kinase.
What is the purpose of purine synthesis?
Biosynthesis of purine ribonucleotides is an essential process that provides the body with important molecules, such as guanine and hypoxanthine. These molecules are essential for many biological processes, including DNA replication, repair, and protein synthesis.
Why is purine important in DNA synthesis?
Pyrimidine is important in DNA synthesis because, when it combines with thymine, it forms a cytosine backbone.
Final words
In conclusion, purine synthesis is a highly regulated process essential for the proper functioning of many biological processes. The production of purine nucleotides is critical for DNA and RNA synthesis and other metabolic pathways and signaling molecules.
The dysregulation of purine metabolism can lead to various disorders, including gout and Lesch-Nyhan syndrome.
In recent years, significant progress has been made in understanding the mechanisms behind purine biosynthesis and its regulation. It has led to the development of novel therapies for diseases associated with purine metabolism, such as gout.
Ongoing research in this area aims further to elucidate the intricate regulatory mechanisms of purine biosynthesis and develop new therapeutic strategies for treating purine metabolism disorders.
The study of purine metabolism remains an active and exciting field with significant potential for future developments.
Understanding the mechanisms behind purine synthesis is critical for developing new treatments for various disorders and expanding our knowledge of the complex processes that govern our biology.
Multiple Choice Questions (MCQ)
1. The scientist who got noble prize twice for work in Biochemistry is
A Korenberg
B Nirenberg
C. Sanger
D Khorana
Explanation: on the sequencing of Primary structure of Protein (Insulin)
2. PRPP glutamyl amidotransferase, the first enzyme uniquely committed to purine synthesis is feedback inhibited by
A CMP
B IMP
C. AMP
D XMP
3. In the biosynthesis of purine nucleotides the AMP feedback regulates
A. Adenylosuccinate synthetase
B HGPRTase
C Adenylosuccinate
D IMP dehydrogenase
4. 6th Carbon of purine skeleton comes from
A Methionine
B 1st carbon carried by Folate
C Betaine
D. Atmospheric CO2
5. Conversion of Formyl Glycinamide ribosyl enzyme uniquely committed to purine synthesis is feedback inhibited by
A Mycophenolic acid
B Diazonoleucine
C 6-mercaptoPurine
D. Azaserine
6. Phosphorylation of Adenosine to AMP catalyzed by
A. Adenosine Kinase
B Adenylosuccinate synthetase
C Deoxycytidine kinase
D Adenylosuccinase
7. In purine synthesis carbon atoms at 4 and 5 position and N at 7th position are contributed by
A. Alanine
B. Glycine
C Threonine
D Glutamine
8. In purine synthesis ring closure in the molecule formyl glycinamide ribosyl-5-phosphate requires the cofactors
A FAD
B. ATP and Mg++
C ADP
D NAD
9. Conversion of IMP to XMP is catalyzed by
A Xanthine-Guanine phosphoribosyl transferase
B Adenine Phosphoribosyl transferase
C Formyl transferase
D. IMP dehydrogenase
10. In Purine nucleus nitrogen atom at 1st position is derived from
A Alanine
B. Aspartate
C Glutamate
D Glycine
11. A common substrate of HGPRTase, APRTase, and PRPP glutamyl amidotransferase is
A Adenosine
B Ribose-5-Phosphate
C Hypoxanthine
D. Phosphoribosyl pyrophosphate
12. Purine biosynthesis is inhibited by
A Chloramphenicol
B Methotrexate
C Tetracyclin
D. Aminopterin
13. Glycine gives _ atoms of purine
A C4, C5 and N9
B C4, C6 and N7
C C2 and C3
D. C4, C5 and N7
14. The cofactors required for the synthesis of “Adenylosuccinate” are
A ATP and Mg++
B. GTP and Mg++
C GDP
D ADP
15. Ring closure of formimidoimidazole carboxamide ribosyl-5-phosphate yields the first purine nucleotide
A XMP
B AMP
C. IMP
D GMP
16. The major determinant of the overall rate of denovo purine nucleotide biosynthesis is the concentration of
A Glycinamide ribosyl-5-Phosphate
B. 5-Phosphoribosyl-1-Pyrophosphate
C 5-Phospho-β-D-Ribosylamine
D FormylGlycinamide ribosyl-5-Phosphate
17. N10-formyl and N5N10-methyl tetrahydrofolate contributes purine carbon atoms at position
A 4 and 6
B 5 and 6
C. 2 and 8
D 4 and 5
18. Uric acid is the catabolic end product of
A. Purines
B Pyrimidines
C Porphyrins
D Pyridoxine
19. Catabolism of Thymidylate gives
A alpha alanine
B alpha amino isobutyrate
C. beta-Amino isobutyrate
D beta alanine
20. The key substance in the synthesis of Purine, phosphoribosyl pyrophosphate is formed by
A D-Ribose
B 5-Phospho-β-D-ribosylamine
C. α-D-ribose-5-Phosphate
D Deoxyribose
Discover more from Biochemistry Den
Subscribe to get the latest posts sent to your email.