Are you looking for information about Stabilizing bonds in protein structure? Proteins are vital macromolecules at both cellular and systemic levels, but they rarely act alone.
Diverse essential molecular processes within a cell are carried out by molecular machines built from a large number of protein components organized by their Protein-Proteins interaction (PPI).
Indeed, these interactions are at the core of the entire interactomics system of any living cell. So, unsurprisingly, aberrant PPIs are based on multiple diseases, such as Creutzfeld-Jacob, Alzheimer’s disease, and cancer.
There are several different types of forces acting on/within a protein molecule. These include:
Stabilizing bonds in protein structure
1. The covalent bonds
These strong bonds result from the sharing of electrons between two atoms. There are four different types.
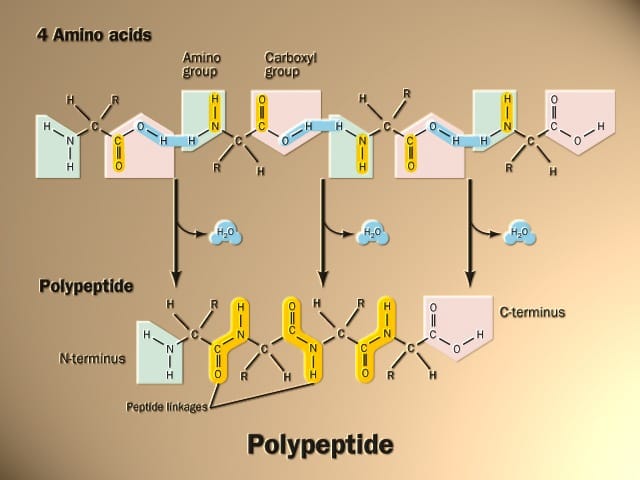
- The peptide bond -CO-NH-bond is responsible for forming the carbon skeleton of the primary structure of proteins. An amide bond is formed between the α carboxylic function of amino acid and another amino acid’s α amino function.
- The carbon-carbon bond -CO-CH is, in fact, in the amino acids making up the protein but plays a role in forming the skeleton.
- The disulfide bridges are found in abundance in proteins and characteristic of globular proteins’ tertiary structure. They are commonly found in fibrous proteins. The bridge stands (optionally) between two cysteines and is broken by the action of mercaptoethanol.
- The desmosine bridges connect four molecules of lysine. Characteristics of elastin are formed by lysyl oxidase.
2. The Non-Covalent Bonds
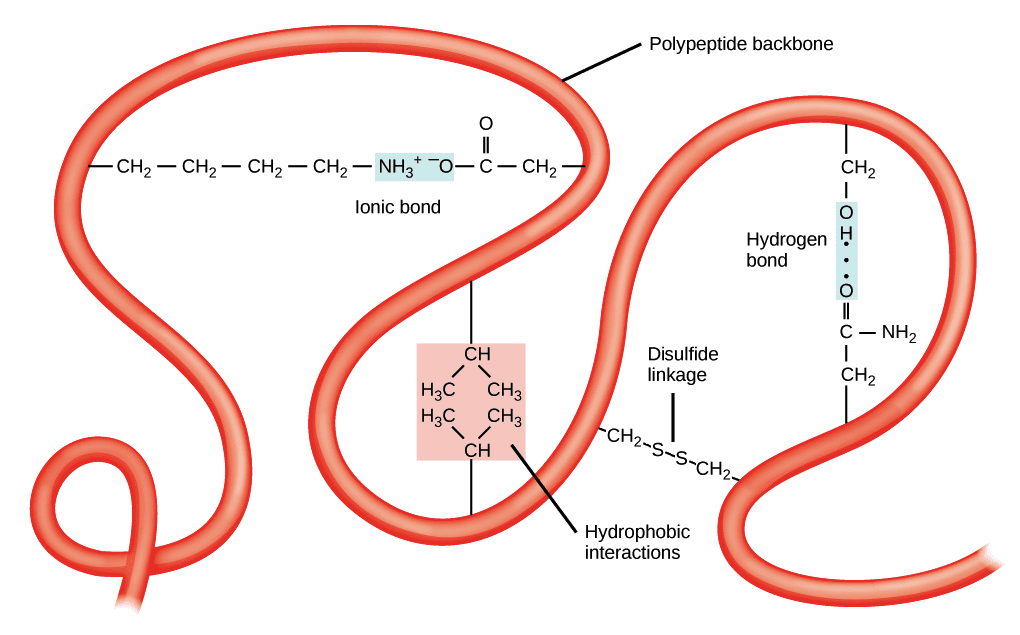
a. The ionic bonds
The ionic bonds are weak bonds resulting from the attraction between two oppositely charged polar groups.
It formed between the carboxyl group and an amino group of an amino acid molecule. These connections can be made within the same chain, the polypeptide folding.
The bonds are the strongest after the covalent bonds. They depend on pH and salt concentrations.
It has specific importance in the relationship between the protein and other molecules.
b.Hydrogen bonds
The hydrogen bonds in protein structure forms between two electronegative atoms roughly shared by one hydrogen atom. An atom is covalently bonded to H is the donor atom. The other atom is the acceptor atom. Generally, we see this connection between the elements of two peptide bonds. They also involve radicals of polar amino acids.
They are weak and unstable, but these defects are compensated by the large number that allows them to have an essential role in maintaining the secondary tertiary structure. Hydrogen bonds gain power when the three atoms are aligned on the same axis.
c.Van der Waals forces
This connection is made to any atom but requires a distance between atoms low: less than 5 angstroms. Each atom has electronic temporary fluctuations making it a transition dipole.
It explains why two atoms, even neutral, can develop a weak interaction, the force of attraction of Van der Waals. Their weakness is offset by their large number, especially when adjusting two macromolecular surfaces.
d.Hydrophobic interactions
They are between two non-polar molecules. In the presence of water, they tend to come together to exclude water.
For example, in the soluble globular proteins, the hydrophobic region (the non-polar groups) is buried within the molecule, excluding water. This phenomenon stabilizes the tertiary structures.
Discover more from Biochemistry Den
Subscribe to get the latest posts sent to your email.